This AI Research Helps Microbiologists to Identify Bacteria
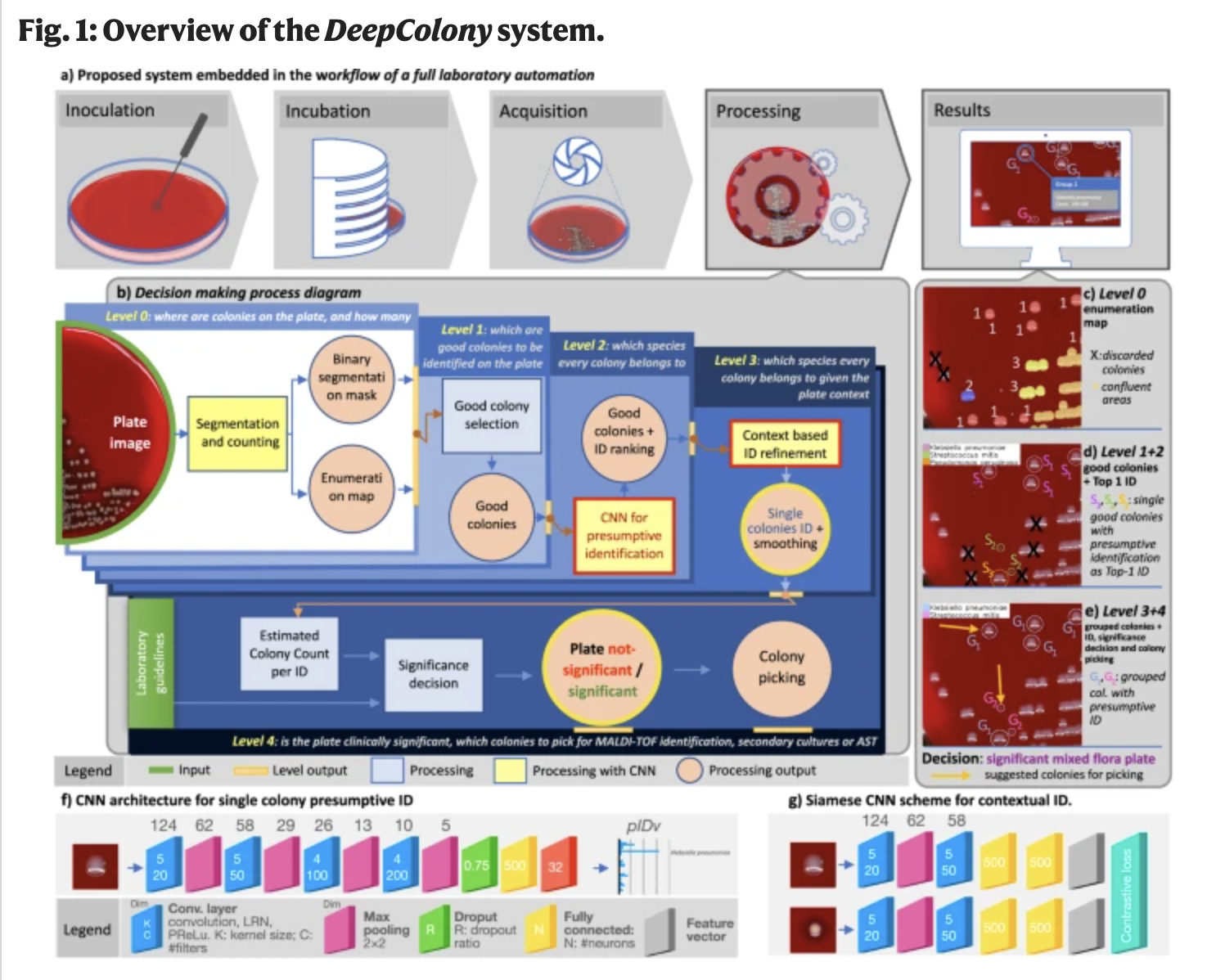
A new AI research presents DeepColony, a comprehensive framework for colony identification and analysis in microbiology laboratories. The system utilizes high-resolution digital scans of cultured plates and employs a hierarchical structure with five levels for colony analysis and identification of bacteria. The levels range from identifying the locations and quantifying the number of colonies on a plate to assessing the clinical significance of the entire plate.
At level 0, DeepColony determines colony locations and quantities, providing essential spatial distribution information. Level 1 identifies isolated colonies for further analysis, considering criteria similar to those used by microbiologists. The core of DeepColony lies in levels 2 to 4, where the system performs initial species identification, refines identification rankings, and assesses the clinical significance of the entire plate.
The system’s architecture involves convolutional neural networks (CNNs) organized in a hierarchical structure. The CNN for single colony identification comprises four convolutional layers and one fully connected layer. DeepColony’s unique approach includes context-based identification, where a Siamese neural network is employed for a non-linear similarity-driven embedding. This embedding, combined with mean-shift clustering, enhances the identification of pathogenic species based on visual data.
The datasets used in the study include colony-level and plate-level data obtained from high-resolution digital scans of cultured plates. The system’s evaluation focused on urine cultures, and the dataset includes a diverse range of organisms.
DeepColony demonstrates the potential to improve the efficiency and quality of routine activities in microbiological laboratories. It can reduce the workload, make coherent decisions aligned with interpretation guidelines, and enhance the role of microbiologists. While the system has limitations, such as difficulty in identifying species in confluent areas, its safety-by-design feature minimizes the impact on result consistency.
In conclusion, DeepColony emerges as a unique framework with the capacity to refine and reinforce the critical role of microbiologists in high-throughput laboratories, offering significant potential for improving decision-making processes in microbiological analyses.
Pragati Jhunjhunwala is a consulting intern at MarktechPost. She is currently pursuing her B.Tech from the Indian Institute of Technology(IIT), Kharagpur. She is a tech enthusiast and has a keen interest in the scope of software and data science applications. She is always reading about the developments in different field of AI and ML.













